Summer is just around the corner, so grab a cold drink, find a shady spot, and catch up on the use of machine learning in cancer research to predict treatment responses, the financial benefits of smoking cessation, and insights into how treatment resistance develops in blood cancer.
For the month of May, the editors of the American Association for Cancer Research’s (AACR) 10 peer-reviewed journals have chosen to highlight these topics and more.
As always, the abstract of each Editors’ Pick can be found below, and the full articles are freely available for a limited time.
Journal: Blood Cancer Discovery
Acquired Multidrug Resistance in AML Is Caused by Low Apoptotic Priming in Relapsed Myeloblasts
In many cancers, mortality is associated with the emergence of relapse with multidrug resistance (MDR). Thus far, the investigation of cancer relapse mechanisms has largely focused on acquired genetic mutations. Using acute myeloid leukemia (AML) patient-derived xenografts (PDX), we systematically elucidated a basis of MDR and identified drug sensitivity in relapsed AML. We derived pharmacologic sensitivity for 22 AML PDX models using dynamic BH3 profiling (DBP), together with genomics and transcriptomics. Using in vivo acquired resistant PDXs, we found that resistance to unrelated, narrowly targeted agents in distinct PDXs was accompanied by broad resistance to drugs with disparate mechanisms. Moreover, baseline mitochondrial apoptotic priming was consistently reduced regardless of the class of drug-inducing selection. By applying DBP, we identified drugs showing effective in vivo activity in resistant models. This study implies evasion of apoptosis drives drug resistance and demonstrates the feasibility of the DBP approach to identify active drugs for patients with relapsed AML.
Significance: Acquired resistance to targeted therapy remains challenging in AML. We found that reduction in mitochondrial priming and common transcriptomic signatures was a conserved mechanism of acquired resistance across different drug classes in vivo. Drugs active in vivo can be identified even in the multidrug resistant state by DBP.
This study was presented during the Blood Cancer Discovery Symposium in March and was featured in a recent blog post.
Journal: Cancer Discovery
Sotorasib Is a Pan-RASG12C Inhibitor Capable of Driving Clinical Response in NRASG12C Cancers
KRASG12C inhibitors, like sotorasib and adagrasib, potently and selectively inhibit KRASG12C through a covalent interaction with the mutant cysteine, driving clinical efficacy in KRASG12C tumors. Because amino acid sequences of the three main RAS isoforms—KRAS, NRAS, and HRAS—are highly similar, we hypothesized that some KRASG12C inhibitors might also target NRASG12C and/or HRASG12C, which are less common but critical oncogenic driver mutations in some tumors. Although some inhibitors, like adagrasib, were highly selective for KRASG12C, others also potently inhibited NRASG12C and/or HRASG12C. Notably, sotorasib was five-fold more potent against NRASG12C compared with KRASG12C or HRASG12C. Structural and reciprocal mutagenesis studies suggested that differences in isoform-specific binding are mediated by a single amino acid: Histidine-95 in KRAS (Leucine-95 in NRAS). A patient with NRASG12C colorectal cancer treated with sotorasib and the anti-EGFR antibody panitumumab achieved a marked tumor response, demonstrating that sotorasib can be clinically effective in NRASG12C-mutated tumors.
Significance: These studies demonstrate that certain KRASG12C inhibitors effectively target all RASG12C mutations and that sotorasib specifically is a potent NRASG12C inhibitor capable of driving clinical responses. These findings have important implications for the treatment of patients with NRASG12C or HRASG12C cancers and could guide design of NRAS or HRAS inhibitors.
This article was featured on the cover of the May issue. A related commentary can be found here.
Journal: Cancer Epidemiology, Biomarkers & Prevention
Disparities in OncotypeDx Testing and Subsequent Chemotherapy Receipt by Geography and Socioeconomic Status
Background: OncotypeDx is a prognostic and predictive genomic assay used in early-stage hormone receptor–positive, HER2− (HR+/HER2−) breast cancer. It is used to inform adjuvant chemotherapy decisions, but not all eligible women receive testing. We aimed to assess variation in testing by demographics and geography, and to determine whether testing was associated with chemotherapy.
Methods: For 1,615 women in the Carolina Breast Cancer Study with HR+/HER2−, Stage I–II tumors, we estimated prevalence differences (PD) and 95% confidence intervals (CI) for receipt of OncotypeDx genomic testing in association with and sociodemographic characteristics. We assessed associations between testing and chemotherapy receipt overall and by race. Finally, we calculated the proportion of eligible women receiving OncotypeDx by county-level rurality, census tract-level socioeconomic status, and Area Health Education Center regions.
Results: 38% (N = 609) of potentially eligible women were tested, with lower testing prevalences in Black (31%; PD, −11%; 95% CI, −16%–6%) and low-income women (24%; PD, −20%; 95% CI, −29% to −11%) relative to non-Black and higher income women. Urban participants were less likely to be tested than rural participants, though this association varied by region. Among women with low genomic risk tumors, tested participants were 29% less likely to receive chemotherapy than untested participants (95% CI, −40% to −17%). Racial differences in chemotherapy were restricted to untested women.
Conclusions: Both individual and area-level socioeconomics predict likelihood of OncotypeDx testing.
Impact: Variable adoption of OncotypeDx by socioeconomics and across geographic settings may contribute to excess chemotherapy among patients with HR+/HER2− cancers.
This article was highlighted in the May issue. A related commentary can be found here.
Journal: Cancer Immunology Research
Machine Learning Links T-cell Function and Spatial Localization to Neoadjuvant Immunotherapy and Clinical Outcome in Pancreatic Cancer
Tumor molecular data sets are becoming increasingly complex, making it nearly impossible for humans alone to effectively analyze them. Here, we demonstrate the power of using machine learning (ML) to analyze a single-cell, spatial, and highly multiplexed proteomic data set from human pancreatic cancer and reveal underlying biological mechanisms that may contribute to clinical outcomes. We designed a multiplex immunohistochemistry antibody panel to compare T-cell functionality and spatial localization in resected tumors from treatment-naïve patients with localized pancreatic ductal adenocarcinoma (PDAC) with resected tumors from a second cohort of patients treated with neoadjuvant agonistic CD40 (anti-CD40) monoclonal antibody therapy. In total, nearly 2.5 million cells from 306 tissue regions collected from 29 patients across both cohorts were assayed, and over 1,000 tumor microenvironment (TME) features were quantified. We then trained ML models to accurately predict anti-CD40 treatment status and disease-free survival (DFS) following anti-CD40 therapy based on TME features. Through downstream interpretation of the ML models’ predictions, we found anti-CD40 therapy reduced canonical aspects of T-cell exhaustion within the TME, as compared with treatment-naïve TMEs. Using automated clustering approaches, we found improved DFS following anti-CD40 therapy correlated with an increased presence of CD44+CD4+ Th1 cells located specifically within cellular neighborhoods characterized by increased T-cell proliferation, antigen experience, and cytotoxicity in immune aggregates. Overall, our results demonstrate the utility of ML in molecular cancer immunology applications, highlight the impact of anti-CD40 therapy on T cells within the TME, and identify potential candidate biomarkers of DFS for anti-CD40–treated patients with PDAC.
This article was featured on the cover of the May issue.
Journal: Cancer Prevention Research
Examining the Association between Abstinence from Smoking and Healthcare Costs among Patients with Cancer
Continuous tobacco use in patients with cancer is linked to substantial healthcare costs due to increased risks and complications, whereas quitting smoking leads to improved treatment outcomes and cost reductions. Addressing the need for empirical evidence on the economic impact of smoking cessation, this study examined the association between smoking cessation and healthcare cost utilization among a sample of 930 patients with cancer treated at The University of Texas MD Anderson Cancer Center’s Tobacco Research and Treatment Program (TRTP). Applying conditional quantile regression and propensity scores to address confounding, our findings revealed that abstinence achieved through the TRTP significantly reduced the median cost during a 3-month period post-quitting by $1,095 [β = −$1,095, P = 0.007, 95% confidence interval (CI), = (−$1,886 to −$304)]. Sensitivity analysis corroborated these conclusions, showing a pronounced cost reduction when outlier data were excluded. The long-term accrued cost savings from smoking cessation could potentially offset the cost of participation in the TRTP program, underscoring its cost effectiveness. An important implication of this study is that by reducing smoking rates, healthcare systems can more efficiently allocate resources, enhance patient health outcomes, and lessen the overall cancer burden.
Prevention Relevance: This study emphasizes the dual impact of smoking cessation programs in patients with cancer: quitting smoking and reducing healthcare costs. It highlights the importance of integrating cessation programs into cancer prevention strategies, ensuring both individual health benefits and broader, system-wide economic efficiencies.
A related commentary can be found here.
Journal: Cancer Research (May 1 issue)
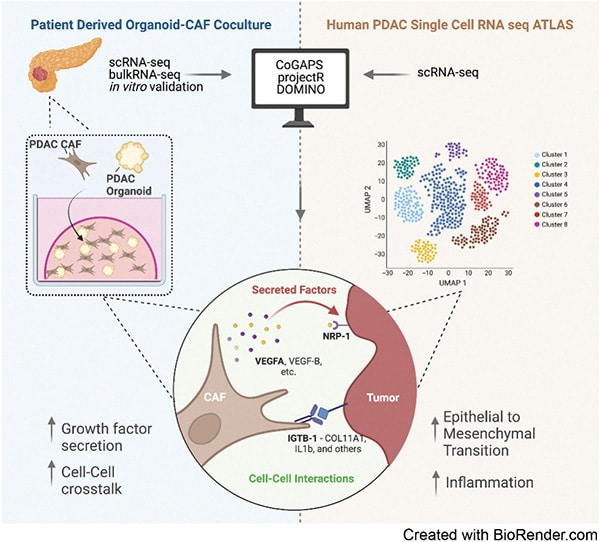
Transfer Learning Reveals Cancer-Associated Fibroblasts Are Associated with Epithelial–Mesenchymal Transition and Inflammation in Cancer Cells in Pancreatic Ductal Adenocarcinoma
Pancreatic ductal adenocarcinoma (PDAC) is an aggressive malignancy characterized by an immunosuppressive tumor microenvironment enriched with cancer-associated fibroblasts (CAF). This study used a convergence approach to identify tumor cell and CAF interactions through the integration of single-cell data from human tumors with human organoid coculture experiments. Analysis of a comprehensive atlas of PDAC single-cell RNA sequencing data indicated that CAF density is associated with increased inflammation and epithelial–mesenchymal transition (EMT) in epithelial cells. Transfer learning using transcriptional data from patient-derived organoid and CAF cocultures provided in silico validation of CAF induction of inflammatory and EMT epithelial cell states. Further experimental validation in cocultures demonstrated integrin beta 1 (ITGB1) and vascular endothelial factor A (VEGFA) interactions with neuropilin-1 mediating CAF-epithelial cell cross-talk. Together, this study introduces transfer learning from human single-cell data to organoid coculture analyses for experimental validation of discoveries of cell–cell cross-talk and identifies fibroblast-mediated regulation of EMT and inflammation.
Significance: Adaptation of transfer learning to relate human single-cell RNA sequencing data to organoid-CAF cocultures facilitates discovery of human pancreatic cancer intercellular interactions and uncovers cross-talk between CAFs and tumor cells through VEGFA and ITGB1.
Journal: Cancer Research (May 15 issue)
Integrin αvβ3 Upregulation in Response to Nutrient Stress Promotes Lung Cancer Cell Metabolic Plasticity
Cancer stem/tumor-initiating cells display stress tolerance and metabolic flexibility to survive in a harsh environment with limited nutrient and oxygen availability. The molecular mechanisms underlying this phenomenon could provide targets to prevent metabolic adaptation and halt cancer progression. Here, we showed in cultured cells and live human surgical biopsies of non–small cell lung cancer that nutrient stress drives the expression of the epithelial cancer stem cell marker integrin αvβ3 via upregulation of the β3 subunit, resulting in a metabolic reprogramming cascade that allows tumor cells to thrive despite a nutrient-limiting environment. Although nutrient deprivation is known to promote acute, yet transient, activation of the stress sensor AMP-activated protein kinase (AMPK), stress-induced αvβ3 expression via Src activation unexpectedly led to secondary and sustained AMPK activation. This resulted in the nuclear localization of peroxisome proliferator-activated receptor-gamma coactivator 1α (PGC1α) and upregulation of glutamine metabolism, the tricarboxylic acid cycle, and oxidative phosphorylation. Pharmacological or genetic targeting of this axis prevented lung cancer cells from evading the effects of nutrient stress, thereby blocking tumor initiation in mice following orthotopic implantation of lung cancer cells. These findings reveal a molecular pathway driven by nutrient stress that results in cancer stem cell reprogramming to promote metabolic flexibility and tumor initiation.
Significance: Upregulation of integrin αvβ3, a cancer stem cell marker, in response to nutrient stress activates sustained AMPK/PGC1α signaling that induces metabolic reprogramming in lung cancer cells to support their survival.
A related commentary can be found here.
Journal: Clinical Cancer Research (May 1 issue)
Phase I/II Study of Combined BCL-xL and MEK Inhibition with Navitoclax and Trametinib in KRAS or NRAS Mutant Advanced Solid Tumors
Purpose: MEK inhibitors (MEKi) lack monotherapy efficacy in most RAS-mutant cancers. BCL-xL is an anti-apoptotic protein identified by a synthetic lethal shRNA screen as a key suppressor of apoptotic response to MEKi.
Patients and Methods: We conducted a dose escalation study (NCT02079740) of the BCL-xL inhibitor navitoclax and MEKi trametinib in patients with RAS-mutant tumors with expansion cohorts for: pancreatic, gynecologic (GYN), non–small cell lung cancer (NSCLC), and other cancers harboring KRAS/NRAS mutations. Paired pretreatment and day 15 tumor biopsies and serial cell-free (cf)DNA were analyzed.
Results: A total of 91 patients initiated treatment, with 38 in dose escalation. Fifty-eight percent had ≥3 prior therapies. A total of 15 patients (17%) had colorectal cancer, 19 (11%) pancreatic, 15 (17%) NSCLC, and 32 (35%) GYN cancers. The recommended phase II dose (RP2D) was established as trametinib 2 mg daily days 1 to 14 and navitoclax 250 mg daily days 1 to 28 of each cycle. Most common adverse events included diarrhea, thrombocytopenia, increased AST/ALT, and acneiform rash. At RP2D, 8 of 49 (16%) evaluable patients achieved partial response (PR). Disease-specific differences in efficacy were noted. In patients with GYN at the RP2D, 7 of 21 (33%) achieved a PR and median duration of response 8.2 months. No PRs occurred in patients with colorectal cancer, NSCLC, or pancreatic cancer. MAPK pathway inhibition was observed in on-treatment tumor biopsies. Reductions in KRAS/NRAS mutation levels in cfDNA correlated with clinical benefit.
Conclusions: Navitoclax in combination with trametinib was tolerable. Durable clinical responses were observed in patients with RAS-mutant GYN cancers, warranting further evaluation in this population.
This article was highlighted in the May 1 issue.
Journal: Clinical Cancer Research (May 15 issue)
Onvansertib in Combination with FOLFIRI and Bevacizumab in Second-Line Treatment of KRAS-Mutant Metastatic Colorectal Cancer: A Phase Ib Clinical Study
Purpose: Onvansertib is a highly specific inhibitor of polo-like kinase 1 (PLK1), with demonstrated safety in solid tumors. We evaluated, preclinically and clinically, the potential of onvansertib in combination with chemotherapy as a therapeutic option for KRAS-mutant colorectal cancer.
Patients and Methods: Preclinical activity of onvansertib was assessed (i) in vitro in KRAS wild-type and -mutant isogenic colorectal cancer cells and (ii) in vivo, in combination with irinotecan, in a KRAS-mutant xenograft model. Clinically, a phase Ib trial was conducted to investigate onvansertib at doses 12, 15, and 18 mg/m2 (days 1–5 and 14–19 of a 28-day cycle) in combination with FOLFIRI/bevacizumab (days 1 and 15) in patients with KRAS-mutant metastatic colorectal cancer who had prior oxaliplatin exposure. Safety, efficacy, and changes in circulating tumor DNA (ctDNA) were assessed.
Results: In preclinical models, onvansertib displayed superior activity in KRAS-mutant than wild-type isogenic colorectal cancer cells and demonstrated potent antitumor activity in combination with irinotecan in vivo. Eighteen patients enrolled in the phase Ib study. Onvansertib recommended phase II dose was established at 15 mg/m2. Grade 3 and 4 adverse events (AE) represented 15% of all treatment-related AEs, with neutropenia being the most common. Partial responses were observed in 44% of patients, with a median duration of response of 9.5 months. Early ctDNA dynamics were predictive of treatment efficacy.
Conclusions: Onvansertib combined with FOLIFRI/bevacizumab exhibited manageable safety and promising efficacy in second-line treatment of patients with KRAS-mutant metastatic colorectal cancer. Further exploration of this combination therapy is ongoing.
This article was highlighted in the May 15 issue. A related commentary can be found here.
Journal: Molecular Cancer Research
Unraveling the Global Proteome and Phosphoproteome of Prostate Cancer Patient-Derived Xenografts
Resistance to androgen-deprivation therapies leads to metastatic castration-resistant prostate cancer (mCRPC) of adenocarcinoma (AdCa) origin that can transform into emergent aggressive variant prostate cancer (AVPC), which has neuroendocrine (NE)-like features. In this work, we used LuCaP patient-derived xenograft (PDX) tumors, clinically relevant models that reflect and retain key features of the tumor from advanced prostate cancer patients. Here we performed proteome and phosphoproteome characterization of 48 LuCaP PDX tumors and identified over 94,000 peptides and 9,700 phosphopeptides corresponding to 7,738 proteins. We compared 15 NE versus 33 AdCa samples, which included six different PDX tumors for each group in biological replicates, and identified 309 unique proteins and 476 unique phosphopeptides that were significantly altered and corresponded to proteins that are known to distinguish these two phenotypes. Assessment of concordance from PDX tumor-matched protein and mRNA revealed increased dissonance in transcriptionally regulated proteins in NE and metabolite interconversion enzymes in AdCa.
Implications: Overall, our study highlights the importance of protein-based identification when compared with RNA and provides a rich resource of new and feasible targets for clinical assay development and in understanding the underlying biology of these tumors.
This article was highlighted in the May issue.
Journal: Molecular Cancer Therapeutics
Design and Evaluation of ZD06519, a Novel Camptothecin Payload for Antibody Drug Conjugates


In recent years, the field of antibody drug conjugates (ADC) has seen a resurgence, largely driven by the clinical benefit observed in patients treated with ADCs incorporating camptothecin-based topoisomerase I inhibitor payloads. Herein, we present the development of a novel camptothecin ZD06519 (FD1), which has been specifically designed for its application as an ADC payload. A panel of camptothecin analogs with different substituents at the C-7 and C-10 positions of the camptothecin core was prepared and tested in vitro. Selected compounds spanning a range of potency and hydrophilicity were elaborated into drug-linkers, conjugated to trastuzumab, and evaluated in vitro and in vivo. ZD06519 was selected on the basis of its favorable properties as a free molecule and as an antibody conjugate, which include moderate free payload potency (∼1 nmol/L), low hydrophobicity, strong bystander activity, robust plasma stability, and high-monomeric ADC content. When conjugated to different antibodies using a clinically validated MC-GGFG–based linker, ZD06519 demonstrated impressive efficacy in multiple cell line–derived xenograft models and noteworthy tolerability in healthy mice, rats, and non-human primates.
This article was highlighted and featured on the cover of the May issue.
Journal: Cancer Research Communications
Overcoming Xenoantigen Immunity to Enable Cellular Tracking and Gene Regulation with Immune-competent “NoGlow” Mice
The ability to temporally regulate gene expression and track labeled cells makes animal models powerful biomedical tools. However, sudden expression of xenobiotic genes [e.g., GFP, luciferase (Luc), or rtTA3] can trigger inadvertent immunity that suppresses foreign protein expression or results in complete rejection of transplanted cells. Germline exposure to foreign antigens somewhat addresses these challenges; however, native fluorescence and bioluminescence abrogates the utility of reporter proteins and highly spatiotemporally restricted expression can lead to suboptimal xenoantigen tolerance. To overcome these unwanted immune responses and enable reliable cell tracking/gene regulation, we developed a novel mouse model that selectively expresses antigen-intact but nonfunctional forms of GFP and Luc, as well as rtTA3, after CRE-mediated recombination. Using tissue-specific CREs, we observed model and sex-based differences in immune tolerance to the encoded xenoantigens, illustrating the obstacles of tolerizing animals to foreign genes and validating the utility of these “NoGlow” mice to dissect mechanisms of central and peripheral tolerance. Critically, tissue unrestricted NoGlow mice possess no detectable background fluorescence or luminescence and exhibit limited adaptive immunity against encoded transgenic xenoantigens after vaccination. Moreover, we demonstrate that NoGlow mice allow tracking and tetracycline-inducible gene regulation of triple-transgenic cells expressing GFP/Luc/rtTA3, in contrast to transgene-negative immune-competent mice that eliminate these cells or prohibit metastatic seeding. Notably, this model enables de novo metastasis from orthotopically implanted, triple-transgenic tumor cells, despite high xenoantigen expression. Altogether, the NoGlow model provides a critical resource for in vivo studies across disciplines, including oncology, developmental biology, infectious disease, autoimmunity, and transplantation.
Significance: Multitolerant NoGlow mice enable tracking and gene manipulation of transplanted tumor cells without immune-mediated rejection, thus providing a platform to investigate novel mechanisms of adaptive immunity related to metastasis, immunotherapy, and tolerance.


